Welcome to UNIO
Last Update: Feb.2024
UNIO NMR App - Overview
The UNIO multi-purpose software suite enables you to conduct unsupervised NMR data analysis for biomolecular 3D structure determination and more..
First released in 2008, UNIO is the ultimate result of more than 15 years of meticulous research performed in order to guarantee accurate, objective and highly automated protein structure determination by NMR.
In 2012 and 2015, UNIO has successively been top-ranked in the NMR community-wide stringent blind target software competition for Critical Assessment of automated Structure Determination by NMR (CASD 1 & 2).
The available UNIO data analysis algorithms and protocols for NMR experiment-driven molecular modelling have been used in thousands of NMR structure determination projects as witnessed by PDB entries.
Academic and industrial licenses have been distributed to more than 1000 research laboratories and structural biology facilities world-wide.
Latest news about UNIO are available in the UNIO blog. Also you can share your UNIO experience with other users in the UNIO forum.
First released in 2008, UNIO is the ultimate result of more than 15 years of meticulous research performed in order to guarantee accurate, objective and highly automated protein structure determination by NMR.
In 2012 and 2015, UNIO has successively been top-ranked in the NMR community-wide stringent blind target software competition for Critical Assessment of automated Structure Determination by NMR (CASD 1 & 2).
The available UNIO data analysis algorithms and protocols for NMR experiment-driven molecular modelling have been used in thousands of NMR structure determination projects as witnessed by PDB entries.
Academic and industrial licenses have been distributed to more than 1000 research laboratories and structural biology facilities world-wide.
Latest news about UNIO are available in the UNIO blog. Also you can share your UNIO experience with other users in the UNIO forum.
UNIO NMR App - Key Algorithms
The UNIO suite includes data analysis routines for all parts of an NMR structure determination process: UNIO assembles the MATCH algorithm for sequence-specific backbone resonance assignment, the ASCAN algorithm for side-chain assignment, the CANDID algorithm for NOE assignment, and the ATNOS algorithm for NMR signal identification. These sophisticated NMR data analysis modules are forming the core of the UNIO software and are controlled by a user-friendly and highly flexible graphical user interface.
UNIO comes with a rich suite of auxiliary modules that are seamlessly integrated into the afore-mentioned principal UNIO analysis components, such as
UNIO comes with a rich suite of auxiliary modules that are seamlessly integrated into the afore-mentioned principal UNIO analysis components, such as
- automated chemical shift referencing
- automated secondary structure prediction
- automated disulfide bond identification
- automated hydrogen bond identification
- automated stereo-specific assignment
- automated order parameter S2 and MD-RMSF identification
- automated identification for optimal RMSD superposition
- use of evolutionary sequence information for accelerated NMR structure determination
- comprehensive validation tools for NMR structures
- easy and flexible molecular conformer visualisation
- NMR restraint and violation inspection
- and more
Automated J-UNIO NMR structure determination
Fully automated NMR structure determination is also provided by UNIO. The standard UNIO data analysis protocol (J-UNIO protocol) requires only a minimal set of six NMR spectra, namely 3 APSY and 3 NOESY, and features high computational efficiency (< 2h for 10-20kDa biomolecules). Triple-resonance NMR experiments can alternatively be used instead of APSY data for obtaining the backbone resonance assignments.
Powerful interactive graphic and text tools facilitate and enhance expert intervention at the structure refinement stage in form of validation, correction and completion of intermediate and final results.
Powerful interactive graphic and text tools facilitate and enhance expert intervention at the structure refinement stage in form of validation, correction and completion of intermediate and final results.
Contact Information

Address
Torsten Herrmann
Research Director CNRS
Institut de Biologie Structurale (IBS)
UMR 5074 CNRS, CEA, UGA
71 avenue des Martyrs, CS 10090
38044 Grenoble cedex 9, France
Get more information
Torsten Herrmann
Research Director CNRS
Institut de Biologie Structurale (IBS)
UMR 5074 CNRS, CEA, UGA
71 avenue des Martyrs, CS 10090
38044 Grenoble cedex 9, France
Get more information
UNIO Manual
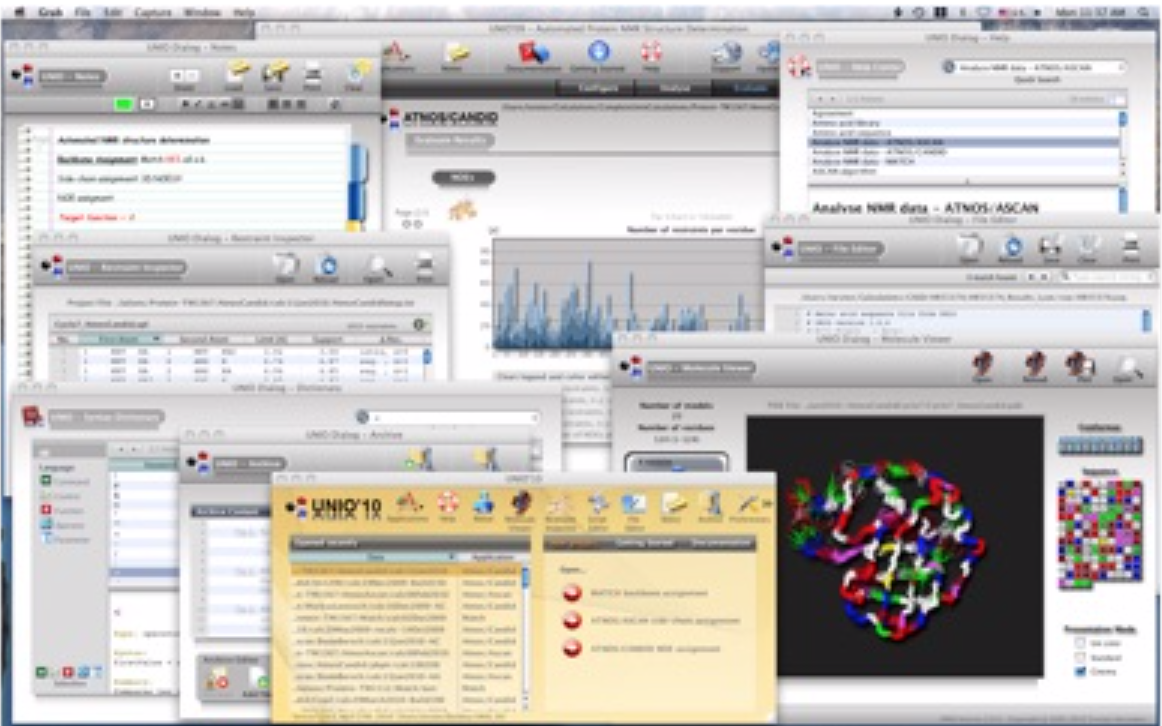
The UNIO manual provides comprehensive documentation with more than 120 pages and over 90 figures.
UNIO References

CASD-NMR 2: Robust and accurate unsupervised analysis of raw NOESY spectra and protein structure determination with UNIO. J. Biomol. NMR 2015, 62(4): 473-480.
APSY-NMR for protein backbone assignment in high-throughput structural biology. J. Biomol. NMR 2015, 61(1): 47-53.
Rapid Proton-Detected NMR Assignment for Proteins with Fast Magic Angle Spinning. J. Am. Chem. Soc. 2014, 136(35): 2489-12497.
The J-UNIO protocol for automated protein structure determination ins solution J. Biol. NMR 2012, 53(4): 341-354.
Comprehensive automation for NMR structure determination of proteins. Methods Mol. Biol. 2012, 831: 429-451.
The complete publication list is available here.